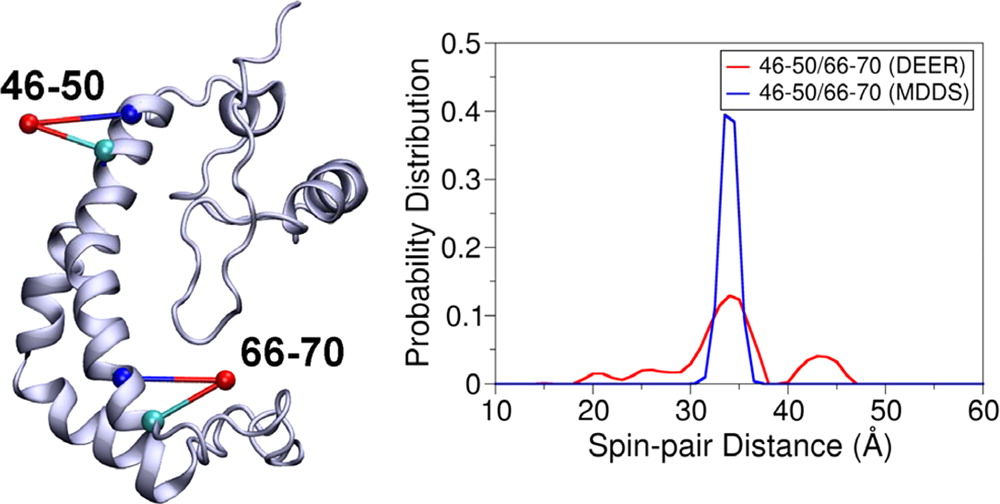
Simulating the Distance Distribution between Spin-Labels Attached to Proteins
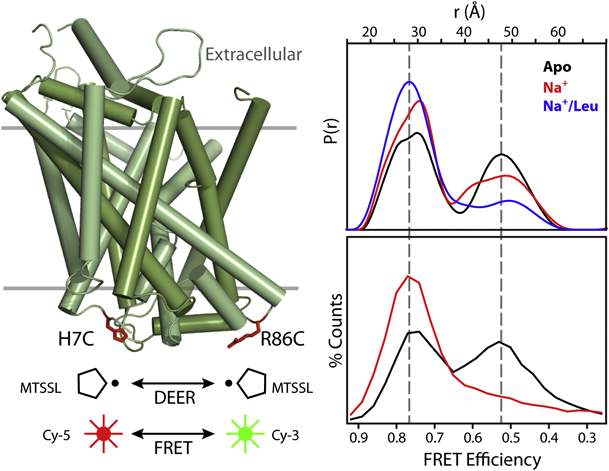
Toward the fourth dimension of membrane protein structure: Insight into dynamics from spin-labeling EPR spectroscopy

The Double‐Histidine Cu2+‐Binding Motif: A Highly Rigid, Site‐Specific Spin Probe for Electron Spin Resonance Distance Measurements - Cunningham - 2015 - Angewandte Chemie - Wiley Online Library

Benchmark Test and Guidelines for DEER/PELDOR Experiments on Nitroxide-Labeled Biomolecules

Simulating the Distance Distribution between Spin-Labels Attached to Proteins
Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance - Physical Chemistry Chemical Physics (RSC Publishing) DOI:10.1039/B614920K

Structural Dynamics of Protein Interactions Using Site-Directed Spin Labeling of Cysteines to Measure Distances and Rotational Dynamics with EPR Spectroscopy
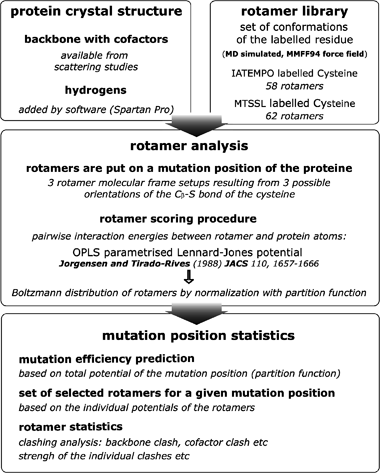
chiLife: An open-source Python package for in silico spin labeling and integrative protein modeling

Site-directed spin labeling-electron paramagnetic resonance spectroscopy in biocatalysis: Enzyme orientation and dynamics in nanoscale confinement - ScienceDirect

Molecular Dynamics Simulations Based on Newly Developed Force Field Parameters for Cu2+ Spin Labels Provide Insights into Double-Histidine-Based Double Electron–Electron Resonance

Simulating the Distance Distribution between Spin-Labels Attached to Proteins

Site-directed spin labeling. The covalent attachment of

Site-directed spin labeling-electron paramagnetic resonance spectroscopy in biocatalysis: Enzyme orientation and dynamics in nanoscale confinement - ScienceDirect









